| Molecular and developmental biology methods: reverse transcription, various PCR and cloning methods, RACE, in situ hybridization |
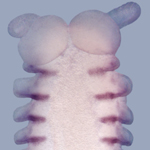
| Molecular and developmental biology methods: reverse transcription, various PCR and cloning methods, RACE, in situ hybridization |

| Phylogenetic analyses using morphological and molecular data sets |
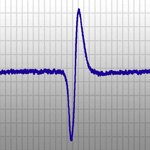
| Electrophysiology (in collaboration with Prof. Paul A. Stevenson) |
| Neuroanatomical methods: Cryotome and Vibratome sectioning and immunolabelling |
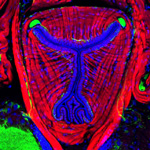
| Neuroanatomical methods: backfills of selected nerve tracts, microinjections of single neurons |
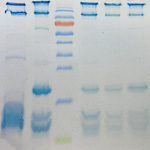
| Proteomics: protein detection by native PAGE and SDS-PAGE, identification of proteins by mass spectrometry (MALDI-TOF, MS/MS), de novo sequencing after protein digestion, and transcriptomics (in collaboration with Prof. Annette G. Beck-Sickinger) |
| Karyotype analyses using Giemsa and fluorescent DNA markers in conjunction with light and confocal microscopy |

| Transmission electron microscopy (TEM) |

| Scanning electron microscopy (SEM) |

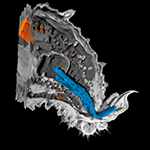
| X-ray micro-computed tomography techniques such as SRµCT, µCT and nanoCT (in collaboration with Dr. Jörg U. Hammel [Synchrotron DESY, Hamburg] and the Department of Diagnostic and Interventional Radiology of the Technical University of Munich) |
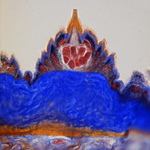
| Histology and light microscopy (LM) |
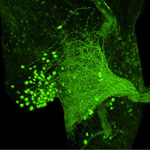
| Histochemistry and immunocytochemistry in conjunction with fluorescence (FM) and confocal laser scanning microscopy (CLSM) |
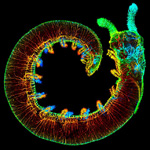
| 3D reconstruction of morphological structures |

| Scientific illustration: ink/pencil drawing, digital imaging, volume rendering, 3D-reconstruction |
| Rearing and breeding of onychophorans in laboratory, both representatives of Peripatopsidae and Peripatidae |
